7.4. LDL decomposition#
LDL decomposition is very similar to Cholesky factorization; however, there are some subtle differences. In this decomposition, a (real) symmetric positive definite matrix \(A\) is decomposed as follows
where \(L\) is a lower triangular matrix, and \(D\) is a diagonal matrix.
Furthermore, We have,
Solving this system, \(l_{ij}\) and \(d_{ii}\) can be identified as follows
See [Allaire, Kaber, Trabelsi, and Allaire, 2008] for the full derivation of this algorithm.
import numpy as np
import pandas as pd
def myLDL(A):
'''
Assuming that the matrix A is symmetric and positive definite,
this function computes a unit lower triangular matrix L, and
a diagonal matrix D, such that A = LDL^t
Parameters
----------
A : numpy array
DESCRIPTION. Matrix A
Returns
-------
L : numpy array
DESCRIPTION. Matrix L from A = LDL^t
D : numpy array
DESCRIPTION. Matrix L from A = LDL^t
'''
n = A.shape[0]
U = np.zeros([n,n], dtype = float)
U[0, 0] = A[0, 0]
U[1:, 0] = A[1:, 0]/U[0, 0]
for j in range(1, n):
U[j, j] = A[j, j]
for k in range(0, j):
U[j, j] = U[j, j] - U[k, k]*U[j, k]*U[j, k]
for i in range(j+1, n):
U[i, j] = A[i, j]
for k in range(0, j):
U[i, j] = U[i, j]- U[k, k]*U[i, k]*U[j, k]
U[i, j] = U[i, j]/U[j, j]
L = np.tril(U,-1) + np.eye(n, dtype=float)
D = np.diag(np.diag(U))
return L, D
function [L, D] = myLDL(A)
%{
Assuming that the matrix A is symmetric and positive definite,
this function computes a unit lower triangular matrix L, and
a diagonal matrix D, such that A = LDL^t
Parameters
----------
L : numpy array
DESCRIPTION. Matrix L from A = LDL^t
D : numpy array
DESCRIPTION. Matrix L from A = LDL^t
%}
n=length(A);
U = zeros(n,n);
U(1,1)=A(1,1);
U(2:n,1)=A(2:n,1)/U(1,1);
for j=2:n
U(j,j)= A(j,j);
for k=1:j-1
U(j,j)=U(j,j)-U(k,k)*U(j,k)*U(j,k);
end
for i=j+1:n
U(i,j)=A(i,j);
for k=1:j-1
U(i,j)=U(i,j)-U(k,k)*U(i,k)*U(j,k);
end
U(i,j)=U(i,j)/U(j,j);
end
%
end
L = tril(U,-1)+eye(n,n);
D = diag(diag(U));
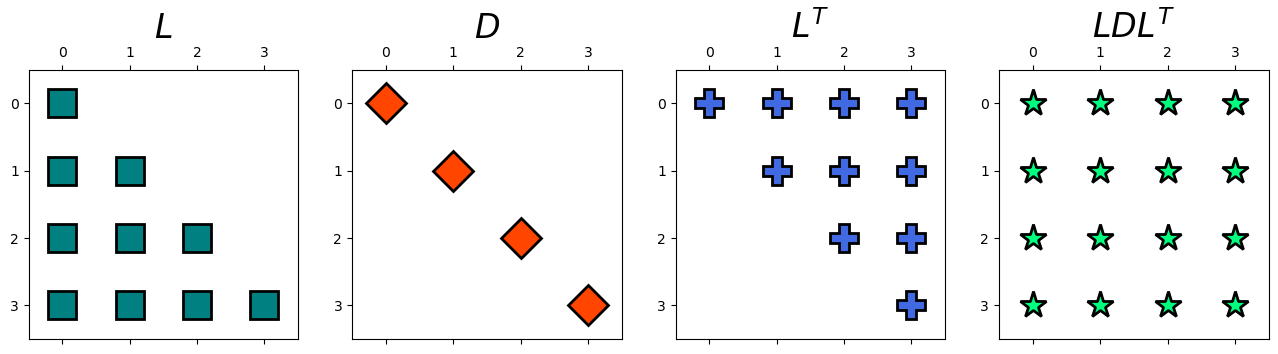
Example: Apply LDL decomposition on the following matrix and identify \(L\), \(D\), and \(L\).
Solution: We have,
import numpy as np
from hd_Matrix_Decomposition import myLDL
from IPython.display import display, Latex
from sympy import init_session, init_printing, Matrix
A = np.array([ [7, 3, -1, 2], [3, 8, 1, -4], [-1, 1, 4, -1], [2, -4, -1, 6] ])
L, D = myLDL(A)
display(Latex(r'L ='), Matrix(np.round(L, 2)))
display(Latex(r'D ='), Matrix(np.round(D, 2)))
display(Latex(r'LD L^T ='), Matrix(np.round(np.matmul(L,np.matmul(D,L.T)), 2)))
hd.matrix_decomp_fig(mats = [L, D, L.T, np.round(np.matmul(L,np.matmul(D,L.T)), 2)],
labels = ['$L$', '$D$','$L^T$', '$LD L^T$'], nrows=1, ncols=4, figsize=(16, 6))
Note that we could get a similar results using function, scipy.linalg.ldl.
import scipy.linalg as linalg
L, D, P = linalg.ldl(A)
display(Latex(r'L ='), Matrix(np.round(L, 2)))
display(Latex(r'D ='), Matrix(np.round(D, 2)))
7.4.1. Solving Linear systems using LDL decomposition#
We can solve the linear system \(Ax=b\) for \(x\) using LDL decomposition. To demonstrate this, we use the following example,
Example: Solve the following linear system using LDL decomposition.
Solution:
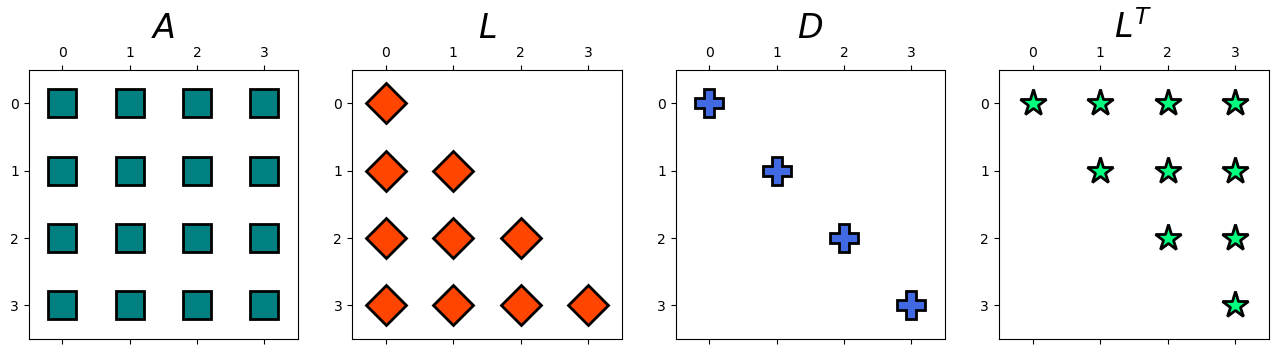
Let \(A=\left[\begin{array}{cccc} 7 & 3 & -1 & 2\\ 3 & 8 & 1 & -4\\ -1 & 1 & 4 & -1\\ 2 & -4 & -1 & 6 \end{array}\right]\) and \(b=\left[\begin{array}{c} 18\\ 6\\ 9\\ 15 \end{array}\right]\). Then, this linear system can be also expressed as follows,
We have,
A = np.array([ [7, 3, -1, 2], [3, 8, 1, -4], [-1, 1, 4, -1], [2, -4, -1, 6] ])
b = np.array([[18],[ 6],[ 9],[15]])
L, D = myLDL(A)
display(Latex(r'L ='), Matrix(np.round(L, 2)))
display(Latex(r'D ='), Matrix(np.round(D, 2)))
Now we can solve the following linear systems instead $\(\begin{cases} Lz = b,\\ Dy = z,\\ B^Tx=y. \end{cases}\)$
hd.matrix_decomp_fig(mats = [A, L, D, L.T], labels = ['$A$', '$L$', '$D$','$L^T$'], nrows=1, ncols=4, figsize=(16, 4))
# solving for z
z = np.linalg.solve(L, b)
display(Latex(r'z ='), Matrix(np.round(z, 2)))
# solving y
y = np.linalg.solve(D, z)
display(Latex(r'y ='), Matrix(np.round(y, 2)))
# solving for x
x = np.linalg.solve(L.T, y)
display(Latex(r'x ='), Matrix(np.round(x, 2)))
Let’s now solve the linear system directly and compare the results.
x_new = linalg.solve(A, b)
display(Latex(r'x ='), Matrix(np.round(x_new, 2)))
